Image: Doi.org-10.7717-peerj.74-Fig-1-full
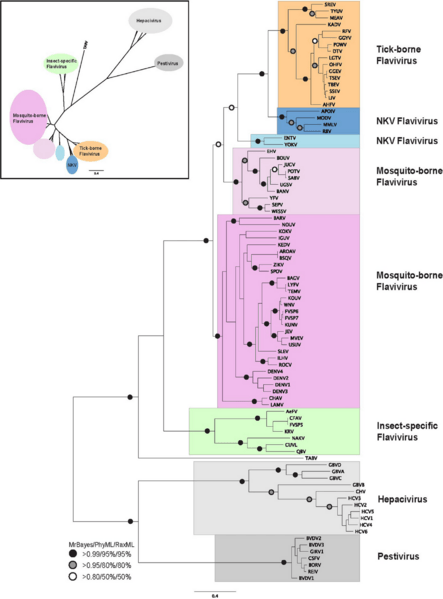
Description: Phylogenetic reconstruction of flaviviridae NS3 protein sequences. The tree shown is the best Bayesian topology. Numerical values at the nodes of the tree (x/y/z) indicate statistical support by MrBayes, PhyML and RAxML (posterior probability, bootstrap and bootstrap, respectively). Values for highly supported nodes have been replaced by symbols, as indicated. Full details and accession numbers for all protein sequences used are given in Table S1. The tree confidently separates the Hepacivirs, Pestivirus, and Flavivirus genera. Within the Flavivirus, TABV and insect-specific species appear basal, whereas Tick-borne species and species with no known vector (NKV) are the most derived. The inset shows the same tree in unrooted star format to better illustrate the relationships and distances between the subgroups.
Title: Doi.org-10.7717-peerj.74-Fig-1-full
Credit: Vlachakis D, Koumandou VL, Kossida S. 2013. A holistic evolutionary and structural study of flaviviridae provides insights into the function and inhibition of HCV helicase. PeerJ 1:e74 https://doi.org/10.7717/peerj.74
Author: Dimitrios Vlachakis, Vassiliki Lila Koumandou, and Sophia Kossida
Usage Terms: Creative Commons Attribution 3.0
License: CC BY 3.0
License Link: https://creativecommons.org/licenses/by/3.0
Attribution Required?: Yes
Image usage
The following page links to this image:

